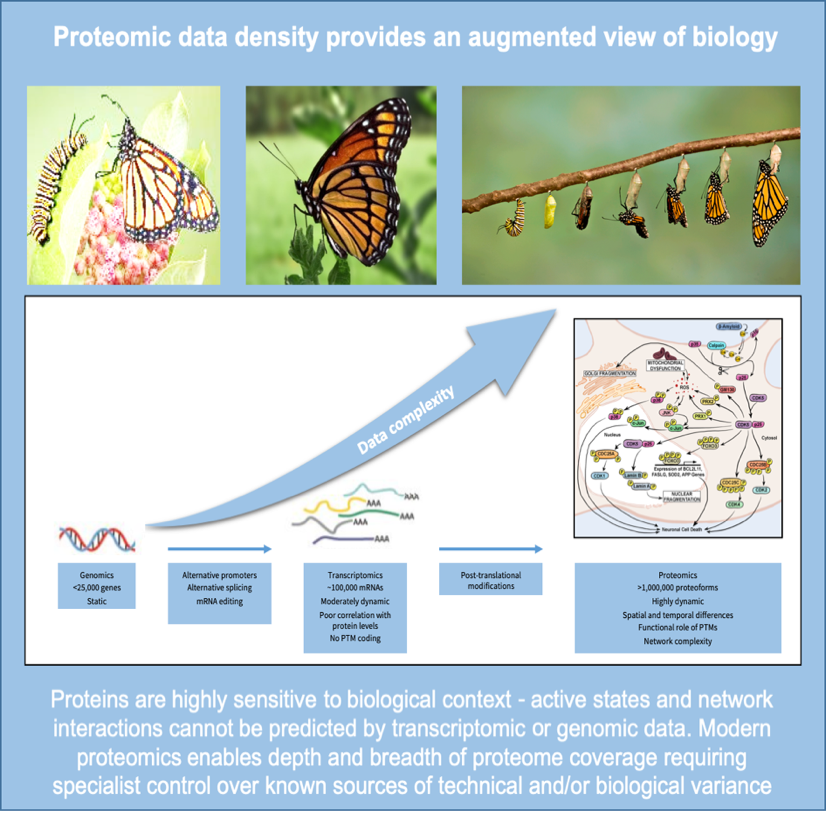
Proteomics data is highly complex and requires powerful but controlled analytical tools. In drug discovery, the majority of drug targets are proteins but changes in form, abundance and function seldom predicted by genome. Combining proteomics with other ‘omics datasets may better enable early diagnosis and personalised treatment.
The past decade has seen a surge in the generation of big genomics datasets triggering the evolution of machine learning (ML) and artificial intelligence (AI) businesses to mine them, usually in combination with clinical information, for actionable biology. This approach is working at the operational level, but outputs mostly remain intangible due to the low predictability of a static genome in a majority of disease settings, and the reliance on public datasets with high variation in quality and lack of cross-study balance. Whilst historically, generation of proteomic datasets of the same order has been challenged by technological and sample-related issues, modern methods, particularly with higher-plexing TMTpro reagents provide robust quantitative analysis of hundreds of thousands of protein isoforms in large cohorts.
Developing functional informatics in healthcare
Interpreting nucleic acid sequences to determine what is happening in a dynamic biological environment is overly simplistic and misses the significant environmental context of health and disease. Regulation of DNA integrity, its replication and translation, and all the functional transactions that make a cell live or die are determined by how proteins interact with each other and their many substrates. We now understand that a single gene can lead to multiple protein products which themselves are modified depending on the environment of the cell at any given time. Changes include post-translational modification of amino acids with labile groups such as phosphorylation, acetylation and methylation, or through linkage of other proteins such as ubiquitin to target them for degradation.
Proteome Sciences is a dedicated proteomics service provider generating datasets with the required quality for ML/AI analysis, both alone and in combination with other ‘omics, to identify the key hallmarks of disease leading to new therapies, diagnostics and personalised treatment. This extends beyond mere quantification to now include post-translational modification profiling and analysis of endogenous protein fractions (peptidomics). We offer ML/AI companies the opportunity to generate the highest quality datasets and analytical tools to supplement their multi-omics projects.
Augmented Omics
The next phase of disruptive technology in healthcare will come through new partnerships between innovators in the ML/AI space and high-quality data generators.
