| Application | Service | Description |
|---|---|---|
| Protein Degrader/PROTAC Assays |
|
Advantages
- Detection and quantification of 10,000+ proteins in 10 to 100s of cell lysate samples
- Can determine all or part of
- the degrader efficiency in respect of the presence of the degraded protein after reaction,
- the presence of degraded peptides arising from the degradation
- the ubiquitylation intermediate
- the effect of protein loss on downstream signalling pathways
- the specificity of the degrader to a single selected protein or cross reactivity of other proteins in the sample proteome
- monitor off target / adverse effects
- Includes full computational proteomic analysis and bioinformatics covering extensive data interpretation, including pathway analysis and biological relevance
Targeted protein degradation through the use of molecular glues and PROTAC’s (proteolysis targeting chimera) is a rapidly evolving field using small molecules to promote selective tagging of target proteins and directing them for proteosomal or lysosomal degradation. Whilst the approach can be highly selective, it is important to monitor the level of protein degradation and look for non-specific protein targeting.
We offer analytical services to support targeted protein degradation drugs at multiple levels (Figure 1).
Peptidomics - identify & measure proteosomal/lysosomal peptide fragments
Proteomics - confirm loss of target protein, assess evidence of cross-reactivity, monitor wider changes in cell signalling and biological processes
Enrichment Proteomics - monitor levels of pro-degradation tagging (Ubiquitin, LC3B etc)
Any of these workflows can be run as single projects, e.g. to only monitor changes in the total proteome before and after Degrader action.
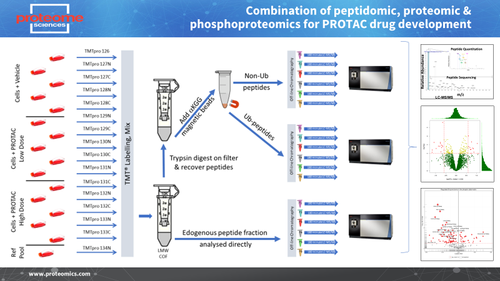
Figure 1 - Three-level analysis of targeted protein degradation using sequential sample preparation and mass spectrometry with TMT and TMTpro reagents.
| What’s included |
|
| Material required |
|
| Typical turnaround time |
|
