| Application | Service | Description |
|---|---|---|
| Routine Biomarker Assays |
Label Free Peptidomics
|
|
| PROTAC/Molecular Glue |
Label Free Peptidomics
|
|
|
Peptidomics MHC I and MHC II Immunopeptidomics |
Label Free Peptidomics
|
Applications for MHC I/II analysis
|
Advantages
- Detection and quantification of non-tryptic or endogenous peptides in 1 to 100s of fluid and tissue samples
- Human, rodent, monkey, pig and others all covered
- Compatibility with a wide variety of sample matrices, including tissue, serum, cell lysates, CSF, exosomes, etc.
- Ability to enrich for specific peptide subsets,
- Includes full computational proteomic analysis and bioinformatics covering extensive data interpretation, including pathway analysis and biological relevance
Peptidomics is the analysis of endogenously created fragments of proteins through the action of targeted or random protease activity. The role of truncated protein fragments in the evolution of many diseases is increasingly recognised and, whilst they are particularly challenging to analyse, they offer novel insights that can deliver new therapeutic targets and biomarkers.
With this increased interest in endogenous protein processing by targeted degraders (PROTAC, molecular glues) and as part of the innate immune response (MHC presentation of tumour antigens) there is a renewed interest in the analysis of the low molecular-weight peptidome.
We have developed specific peptidomics workflows that combine protein-level labelling with TMT and TMTpro , isolation of small endogenous peptides with ultrafiltration, followed by no-digest mass spectrometry (Figure 1). This can be used for analysis of cells, culture supernatants, body fluids (serum/plasma, CSF, urine) or tissues.
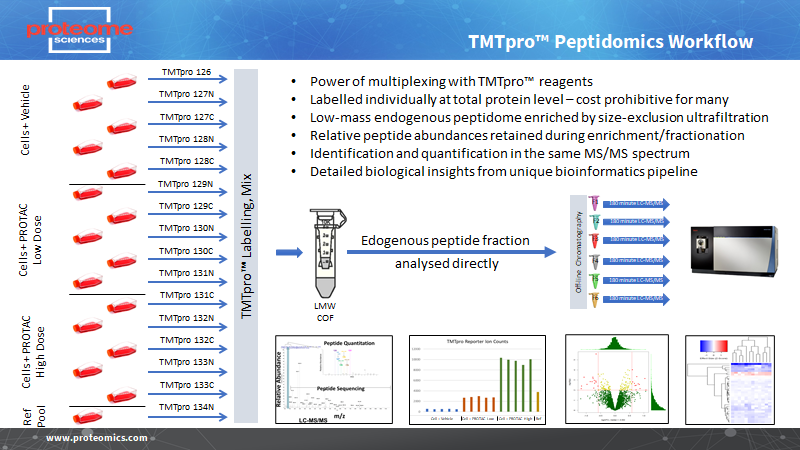
We can also combine peptidomics with the TMTcalibrator workflow where endogenous peptides from tissues or synthetic reference peptides can be labelled and combined with equivalent peptides in body fluids, providing benefits of signal amplification and greater sensitivity.
Featured peptidomics publication:
“An Integrated Workflow for Multiplex CSF Proteomics and Peptidomics Identification of Candidate Cerebrospinal Fluid Biomarkers of Alzheimer’s Disease.” This paper, co-authored by Proteome Sciences and partners at University of Gothenburg, presents a workflow for simultaneous analysis of endogenous peptides and proteins in CSF using multiplex isobaric labelling for quantification. By using this workflow, we could compare the abundances of 437 endogenous peptides and 374 proteins in 100 μL of CSF. A pilot study in a small clinical material demonstrated the potential use of this method to identify biomarker candidates of AD.
| What’s included |
|
| Material required |
|
| Typical turnaround time |
|
